My Publications
Peer-Reviewed Journals
Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4
Nature Genetics, 46, 427–429 (2014)
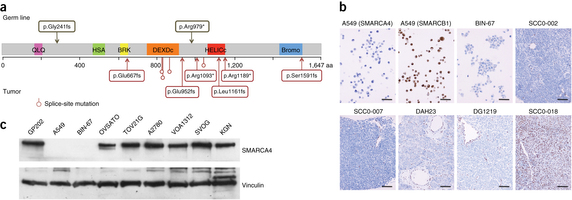 Small cell carcinoma of the ovary of hypercalcemic type (SCCOHT) is an extremely rare, aggressive cancer affecting children and young women. We identified germline and somatic inactivating mutations in the SWI/SNF chromatin-remodeling gene SMARCA4 in 69% (9/13) of SCCOHT cases in addition to SMARCA4 protein loss in 82% (14/17) of SCCOHT tumors but in only 0.4% (2/485) of other primary ovarian tumors. These data implicate SMARCA4 in SCCOHT oncogenesis.
Small cell carcinoma of the ovary of hypercalcemic type (SCCOHT) is an extremely rare, aggressive cancer affecting children and young women. We identified germline and somatic inactivating mutations in the SWI/SNF chromatin-remodeling gene SMARCA4 in 69% (9/13) of SCCOHT cases in addition to SMARCA4 protein loss in 82% (14/17) of SCCOHT tumors but in only 0.4% (2/485) of other primary ovarian tumors. These data implicate SMARCA4 in SCCOHT oncogenesis.
Yang Y, Engkvist O, Llinàs A, Chen H. Beyond Size, Ionization State, and Lipophilicity: Influence of Molecular Topology on Absorption, Distribution, Metabolism, Excretion, and Toxicity for Druglike Compounds
Journal of Medicinal Chemistry, 55 (8), 3667–3677 (2012)
Recommended by F1000Prime: f1000.com/prime/14263965
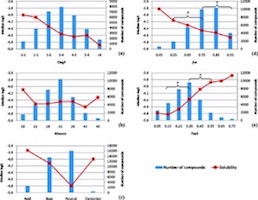 The absorption, distribution, metabolism, excretion, and toxicity (ADMET) of a compound is dependent on physicochemical properties such as molecular size, lipophilicity, and ionization state. However, much less is known regarding the relationship between ADMET and the molecular topology. In this study two descriptors related to the molecular topology have been investigated, the fraction of the molecular framework (fMF) and the fraction of sp3-hybridized carbon atoms (Fsp3). fMF and Fsp3, together with standard physicochemical properties (molecular size, ionization state, and lipophilicity), were analyzed for a set of ADMET assays. It is shown that aqueous solubility, Caco-2 permeability, plasma protein binding, human ether-a-go-go-related potassium channel protein inhibition, and CYP3A4 (CYP = cytochrome P450) inhibition are influenced by the molecular topology. These findings are in most cases independent of the already well-established relationships between the properties and molecular size, lipophilicity, and ionization state.
The absorption, distribution, metabolism, excretion, and toxicity (ADMET) of a compound is dependent on physicochemical properties such as molecular size, lipophilicity, and ionization state. However, much less is known regarding the relationship between ADMET and the molecular topology. In this study two descriptors related to the molecular topology have been investigated, the fraction of the molecular framework (fMF) and the fraction of sp3-hybridized carbon atoms (Fsp3). fMF and Fsp3, together with standard physicochemical properties (molecular size, ionization state, and lipophilicity), were analyzed for a set of ADMET assays. It is shown that aqueous solubility, Caco-2 permeability, plasma protein binding, human ether-a-go-go-related potassium channel protein inhibition, and CYP3A4 (CYP = cytochrome P450) inhibition are influenced by the molecular topology. These findings are in most cases independent of the already well-established relationships between the properties and molecular size, lipophilicity, and ionization state.
Molecular Topology Analysis of the Differences between Drugs, Clinical Candidate Compounds, and Bioactive Molecules
Journal of Chemical Information and Modelling, 50 (12), 2141–2150 (2012)
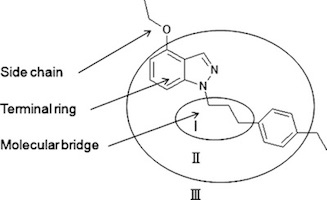 A new method to decompose molecules is proposed and used to analyze drugs, clinical candidate compounds and bioactive molecules. The method classifies a set of molecules into a few well-defined classes based on their molecular framework. It is then possible to use these classes to investigate differences between drugs, clinical candidates and bioactive molecules. The analysis shows that in comparison with clinical candidates and bioactive compounds, drugs have a higher fraction of compounds with only one ring system. This conclusion is still valid after correcting for lipophilicity (ClogP) and molecular size, as well as any potential protein target bias in the data sets. Furthermore the molecular bridge part of compounds in the drug set has on average fewer ring systems than molecules from the other sets. The ring system complexity (RSC) was also investigated and for most topological classes drugs have a lower RSC than the clinical candidates and bioactive molecules. Hence, this study highlights differences in topology between drugs, clinical candidate compounds and bioactive molecules.
A new method to decompose molecules is proposed and used to analyze drugs, clinical candidate compounds and bioactive molecules. The method classifies a set of molecules into a few well-defined classes based on their molecular framework. It is then possible to use these classes to investigate differences between drugs, clinical candidates and bioactive molecules. The analysis shows that in comparison with clinical candidates and bioactive compounds, drugs have a higher fraction of compounds with only one ring system. This conclusion is still valid after correcting for lipophilicity (ClogP) and molecular size, as well as any potential protein target bias in the data sets. Furthermore the molecular bridge part of compounds in the drug set has on average fewer ring systems than molecules from the other sets. The ring system complexity (RSC) was also investigated and for most topological classes drugs have a lower RSC than the clinical candidates and bioactive molecules. Hence, this study highlights differences in topology between drugs, clinical candidate compounds and bioactive molecules.
Yang Y, Chen H, Nilsson I, Muresan S, Engkvist O. Investigation of the Relationship between Topology and Selectivity for Druglike Molecules
Journal of Medicinal Chemistry, 53 (21), 7709–7714 (2010)
Recommended by F1000Prime: f1000.com/prime/5727956
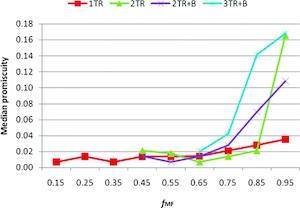 There is a strong interest in drug discovery and development to advance the understanding of pharmacological promiscuity. Improved understanding of how a molecular structure is related to promiscuity could help to reduce the attrition of compounds in the drug discovery process. For this purpose, a descriptor is introduced that describes the structural complexity of a compound based on the size of its molecular framework (MF) in relation to its overall size. It is defined as the fraction of the size of the molecular framework versus the size of the whole molecule (fMF). It is demonstrated that promiscuity correlates with fMF for large fMF values. The observed correlation is not due to lipophilicity. To provide further explanation of this observation, it was found that the number of terminal ring systems in a compound is correlated with promiscuity. The analysis presented here might help medicinal chemists to improve the selectivity for compounds in drug discovery projects.
There is a strong interest in drug discovery and development to advance the understanding of pharmacological promiscuity. Improved understanding of how a molecular structure is related to promiscuity could help to reduce the attrition of compounds in the drug discovery process. For this purpose, a descriptor is introduced that describes the structural complexity of a compound based on the size of its molecular framework (MF) in relation to its overall size. It is defined as the fraction of the size of the molecular framework versus the size of the whole molecule (fMF). It is demonstrated that promiscuity correlates with fMF for large fMF values. The observed correlation is not due to lipophilicity. To provide further explanation of this observation, it was found that the number of terminal ring systems in a compound is correlated with promiscuity. The analysis presented here might help medicinal chemists to improve the selectivity for compounds in drug discovery projects.
‘tieredScreen’ – Layered Virtual Screening Tool for the Identification of Novel Estrogen Receptor Alpha Modulators
Molecular Informatics, 29(5), 421-430 (2010)
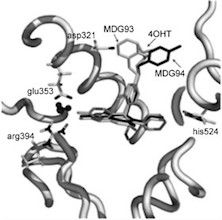 A novel tiered Structure-Based (SB) Virtual Screening (VS) workflow called tieredScreen was designed and implemented. The automated protocol utilises diverse computational tools in a synergistic manner to reduce false positives and increase the likelihood of converging on putative active molecules. The performance of the novel VS workflow was validated using the Directory of Useful Decoys (DUD) Estrogen Receptor α (ERα) antagonist dataset, and successfully deployed for the identification of novel antagonists of ERα from a screening collection of ca. 160 000 commercially available compounds. As well as yielding nanomolar (nM) active ligands identified previously through a docking only protocol, from a selection of eight virtual hits suggested by tieredScreen, four novel nM ERα binding chemotypes were identified and biologically validated - demonstrating the applicability of a tiered intervention for virtual screening.
A novel tiered Structure-Based (SB) Virtual Screening (VS) workflow called tieredScreen was designed and implemented. The automated protocol utilises diverse computational tools in a synergistic manner to reduce false positives and increase the likelihood of converging on putative active molecules. The performance of the novel VS workflow was validated using the Directory of Useful Decoys (DUD) Estrogen Receptor α (ERα) antagonist dataset, and successfully deployed for the identification of novel antagonists of ERα from a screening collection of ca. 160 000 commercially available compounds. As well as yielding nanomolar (nM) active ligands identified previously through a docking only protocol, from a selection of eight virtual hits suggested by tieredScreen, four novel nM ERα binding chemotypes were identified and biologically validated - demonstrating the applicability of a tiered intervention for virtual screening.
Virtual screening of the estrogen receptor
Expert Opinion on Drug Discovery, 3(8), 853-866 (2008)
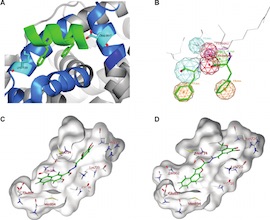 A novel tiered Structure-Based (SB) Virtual Screening (VS) workflow called tieredScreen was designed and implemented. The automated protocol utilises diverse computational tools in a synergistic manner to reduce false positives and increase the likelihood of converging on putative active molecules. The performance of the novel VS workflow was validated using the Directory of Useful Decoys (DUD) Estrogen Receptor α (ERα) antagonist dataset, and successfully deployed for the identification of novel antagonists of ERα from a screening collection of ca. 160 000 commercially available compounds. As well as yielding nanomolar (nM) active ligands identified previously through a docking only protocol, from a selection of eight virtual hits suggested by tieredScreen, four novel nM ERα binding chemotypes were identified and biologically validated - demonstrating the applicability of a tiered intervention for virtual screening.
A novel tiered Structure-Based (SB) Virtual Screening (VS) workflow called tieredScreen was designed and implemented. The automated protocol utilises diverse computational tools in a synergistic manner to reduce false positives and increase the likelihood of converging on putative active molecules. The performance of the novel VS workflow was validated using the Directory of Useful Decoys (DUD) Estrogen Receptor α (ERα) antagonist dataset, and successfully deployed for the identification of novel antagonists of ERα from a screening collection of ca. 160 000 commercially available compounds. As well as yielding nanomolar (nM) active ligands identified previously through a docking only protocol, from a selection of eight virtual hits suggested by tieredScreen, four novel nM ERα binding chemotypes were identified and biologically validated - demonstrating the applicability of a tiered intervention for virtual screening.